#!conda install pyarrow
#!pip install pydicom kornia opencv-python scikit-image nbdevChest X-ray model
To use fastai.medical.imaging you’ll need to:
conda install pyarrow
pip install pydicom kornia opencv-python scikit-imageTo run this tutorial on Google Colab, you’ll need to uncomment the following two lines and run the cell:
from fastai.basics import *
from fastai.callback.all import *
from fastai.vision.all import *
from fastai.medical.imaging import *
import pydicom
import pandas as pdDownload and import of X-ray DICOM files
First, we will use the untar_data function to download the siim_small folder containing a subset (250 DICOM files, ~30MB) of the SIIM-ACR Pneumothorax Segmentation [1] dataset. The downloaded siim_small folder will be stored in your ~/.fastai/data/ directory. The variable pneumothorax-source will store the absolute path to the siim_small folder as soon as the download is complete.
pneumothorax_source = untar_data(URLs.SIIM_SMALL)The siim_small folder has the following directory/file structure:
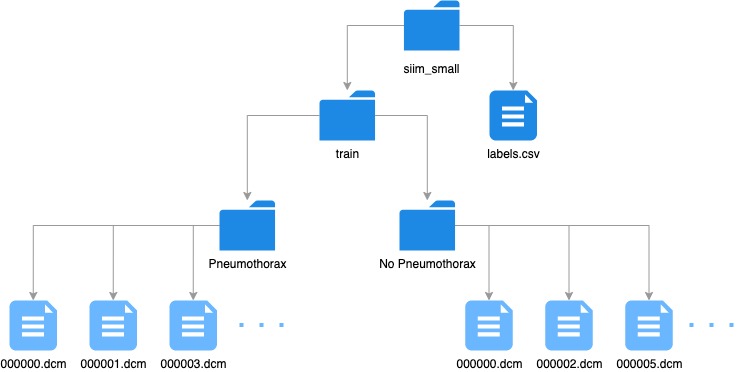
What are DICOMs?
DICOM(Digital Imaging and COmmunications in Medicine) is the de-facto standard that establishes rules that allow medical images(X-Ray, MRI, CT) and associated information to be exchanged between imaging equipment from different vendors, computers, and hospitals. The DICOM format provides a suitable means that meets health infomation exchange (HIE) standards for transmision of health related data among facilites and HL7 standards which is the messaging standard that enables clinical applications to exchange data
DICOM files typically have a .dcm extension and provides a means of storing data in separate ‘tags’ such as patient information as well as image/pixel data. A DICOM file consists of a header and image data sets packed into a single file. By extracting data from these tags one can access important information regarding the patient demographics, study parameters, etc.
16 bit DICOM images have values ranging from -32768 to 32768 while 8-bit greyscale images store values from 0 to 255. The value ranges in DICOM images are useful as they correlate with the Hounsfield Scale which is a quantitative scale for describing radiodensity
Plotting the DICOM data
To analyze our dataset, we load the paths to the DICOM files with the get_dicom_files function. When calling the function, we append train/ to the pneumothorax_source path to choose the folder where the DICOM files are located. We store the path to each DICOM file in the items list.
items = get_dicom_files(pneumothorax_source/f"train/")Next, we split the items list into a train trn and validation val list using the RandomSplitter function:
trn,val = RandomSplitter()(items)Pydicom is a python package for parsing DICOM files, making it easier to access the header of the DICOM as well as coverting the raw pixel_data into pythonic structures for easier manipulation. fastai.medical.imaging uses pydicom.dcmread to load the DICOM file.
To plot an X-ray, we can select an entry in the items list and load the DICOM file with dcmread.
patient = 7
xray_sample = items[patient].dcmread()To view the header
xray_sampleDataset.file_meta -------------------------------
(0002, 0000) File Meta Information Group Length UL: 200
(0002, 0001) File Meta Information Version OB: b'\x00\x01'
(0002, 0002) Media Storage SOP Class UID UI: Secondary Capture Image Storage
(0002, 0003) Media Storage SOP Instance UID UI: 1.2.276.0.7230010.3.1.4.8323329.3297.1517875177.149805
(0002, 0010) Transfer Syntax UID UI: JPEG Baseline (Process 1)
(0002, 0012) Implementation Class UID UI: 1.2.276.0.7230010.3.0.3.6.0
(0002, 0013) Implementation Version Name SH: 'OFFIS_DCMTK_360'
-------------------------------------------------
(0008, 0005) Specific Character Set CS: 'ISO_IR 100'
(0008, 0016) SOP Class UID UI: Secondary Capture Image Storage
(0008, 0018) SOP Instance UID UI: 1.2.276.0.7230010.3.1.4.8323329.3297.1517875177.149805
(0008, 0020) Study Date DA: '19010101'
(0008, 0030) Study Time TM: '000000.00'
(0008, 0050) Accession Number SH: ''
(0008, 0060) Modality CS: 'CR'
(0008, 0064) Conversion Type CS: 'WSD'
(0008, 0090) Referring Physician's Name PN: ''
(0008, 103e) Series Description LO: 'view: PA'
(0010, 0010) Patient's Name PN: '6633c659-9249-443e-9851-b83782d1b111'
(0010, 0020) Patient ID LO: '6633c659-9249-443e-9851-b83782d1b111'
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: 'M'
(0010, 1010) Patient's Age AS: '21'
(0018, 0015) Body Part Examined CS: 'CHEST'
(0018, 5101) View Position CS: 'PA'
(0020, 000d) Study Instance UID UI: 1.2.276.0.7230010.3.1.2.8323329.3297.1517875177.149804
(0020, 000e) Series Instance UID UI: 1.2.276.0.7230010.3.1.3.8323329.3297.1517875177.149803
(0020, 0010) Study ID SH: ''
(0020, 0011) Series Number IS: "1"
(0020, 0013) Instance Number IS: "1"
(0020, 0020) Patient Orientation CS: ''
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0010) Rows US: 1024
(0028, 0011) Columns US: 1024
(0028, 0030) Pixel Spacing DS: [0.14300000000000002, 0.14300000000000002]
(0028, 0100) Bits Allocated US: 8
(0028, 0101) Bits Stored US: 8
(0028, 0102) High Bit US: 7
(0028, 0103) Pixel Representation US: 0
(0028, 2110) Lossy Image Compression CS: '01'
(0028, 2114) Lossy Image Compression Method CS: 'ISO_10918_1'
(7fe0, 0010) Pixel Data OB: Array of 161452 elementsExplanation of each element is beyond the scope of this tutorial but this site has some excellent information about each of the entries
Some key pointers on the tag information above:
- Pixel Data (7fe0 0010) - This is where the raw pixel data is stored. The order of pixels encoded for each image plane is left to right, top to bottom, i.e., the upper left pixel (labeled 1,1) is encoded first
- Photometric Interpretation (0028, 0004) - also known as color space. In this case it is
MONOCHROME2where pixel data is represented as a single monochrome image plane where low values=dark, high values=bright. If the colorspace wasMONOCHROMEthen the low values=bright and high values=dark info. - Samples per Pixel (0028, 0002) - This should be 1 as this image is monochrome. This value would be 3 if the color space was RGB for example
- Bits Stored (0028 0101) - Number of bits stored for each pixel sample. Typical 8 bit images have a pixel range between
0and255 - Pixel Represenation(0028 0103) - can either be unsigned(0) or signed(1)
- Lossy Image Compression (0028 2110) -
00image has not been subjected to lossy compression.01image has been subjected to lossy compression. - Lossy Image Compression Method (0028 2114) - states the type of lossy compression used (in this case
ISO_10918_1represents JPEG Lossy Compression) - Pixel Data (7fe0, 0010) - Array of 161452 elements represents the image pixel data that pydicom uses to convert the pixel data into an image.
What does PixelData look like?
xray_sample.PixelData[:200]b'\xfe\xff\x00\xe0\x00\x00\x00\x00\xfe\xff\x00\xe0\x9cv\x02\x00\xff\xd8\xff\xdb\x00C\x00\x03\x02\x02\x02\x02\x02\x03\x02\x02\x02\x03\x03\x03\x03\x04\x06\x04\x04\x04\x04\x04\x08\x06\x06\x05\x06\t\x08\n\n\t\x08\t\t\n\x0c\x0f\x0c\n\x0b\x0e\x0b\t\t\n\x11\n\x0e\x0f\x10\x10\x11\x10\n\x0c\x12\x13\x12\x10\x13\x0f\x10\x10\x10\xff\xc0\x00\x0b\x08\x04\x00\x04\x00\x01\x01\x11\x00\xff\xc4\x00\x1d\x00\x00\x02\x03\x01\x01\x01\x01\x01\x00\x00\x00\x00\x00\x00\x00\x00\x04\x05\x02\x03\x06\x00\x01\x07\x08\t\xff\xc4\x00R\x10\x00\x02\x01\x03\x03\x02\x04\x03\x06\x05\x04\x00\x04\x01\x02\x17\x01\x02\x11\x00\x03!\x04\x121\x05A\x13"Qa\x06q\x81\x142\x91\xa1\xb1\xf0#B\xc1\xd1\xe1\x07\x15R\xf1\x16$3br\x08%4C&cs\x82\x92\xa2'Because of the complexity in interpreting PixelData, pydicom provides an easy way to get it in a convenient form: pixel_array which returns a numpy.ndarray containing the pixel data:
xray_sample.pixel_array, xray_sample.pixel_array.shape(array([[ 0, 0, 0, ..., 13, 13, 5],
[ 0, 0, 0, ..., 13, 13, 5],
[ 0, 0, 0, ..., 13, 12, 5],
...,
[ 0, 0, 0, ..., 5, 3, 0],
[ 0, 0, 0, ..., 6, 4, 0],
[ 0, 0, 0, ..., 8, 5, 0]], dtype=uint8),
(1024, 1024))You can then use the show function to view the image
xray_sample.show()You can also conveniently create a dataframe with all the tag information as columns for all the images in a dataset by using from_dicoms
dicom_dataframe = pd.DataFrame.from_dicoms(items)
dicom_dataframe[:5]| SpecificCharacterSet | SOPClassUID | SOPInstanceUID | StudyDate | StudyTime | AccessionNumber | Modality | ConversionType | ReferringPhysicianName | SeriesDescription | ... | LossyImageCompression | LossyImageCompressionMethod | fname | MultiPixelSpacing | PixelSpacing1 | img_min | img_max | img_mean | img_std | img_pct_window | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ISO_IR 100 | 1.2.840.10008.5.1.4.1.1.7 | 1.2.276.0.7230010.3.1.4.8323329.6904.1517875201.850819 | 19010101 | 000000.00 | CR | WSD | view: PA | ... | 01 | ISO_10918_1 | C:\Users\tijme\.fastai\data\siim_small\train\No Pneumothorax\000000.dcm | 1 | 0.168 | 0 | 254 | 160.398039 | 53.854885 | 0.087029 | ||
| 1 | ISO_IR 100 | 1.2.840.10008.5.1.4.1.1.7 | 1.2.276.0.7230010.3.1.4.8323329.11028.1517875229.983789 | 19010101 | 000000.00 | CR | WSD | view: PA | ... | 01 | ISO_10918_1 | C:\Users\tijme\.fastai\data\siim_small\train\No Pneumothorax\000002.dcm | 1 | 0.143 | 0 | 250 | 114.524713 | 70.752315 | 0.326269 | ||
| 2 | ISO_IR 100 | 1.2.840.10008.5.1.4.1.1.7 | 1.2.276.0.7230010.3.1.4.8323329.11444.1517875232.977506 | 19010101 | 000000.00 | CR | WSD | view: PA | ... | 01 | ISO_10918_1 | C:\Users\tijme\.fastai\data\siim_small\train\No Pneumothorax\000005.dcm | 1 | 0.143 | 0 | 246 | 132.218334 | 73.023531 | 0.266901 | ||
| 3 | ISO_IR 100 | 1.2.840.10008.5.1.4.1.1.7 | 1.2.276.0.7230010.3.1.4.8323329.32219.1517875159.70802 | 19010101 | 000000.00 | CR | WSD | view: PA | ... | 01 | ISO_10918_1 | C:\Users\tijme\.fastai\data\siim_small\train\No Pneumothorax\000006.dcm | 1 | 0.171 | 0 | 255 | 153.405355 | 59.543063 | 0.144505 | ||
| 4 | ISO_IR 100 | 1.2.840.10008.5.1.4.1.1.7 | 1.2.276.0.7230010.3.1.4.8323329.32395.1517875160.396775 | 19010101 | 000000.00 | CR | WSD | view: PA | ... | 01 | ISO_10918_1 | C:\Users\tijme\.fastai\data\siim_small\train\No Pneumothorax\000007.dcm | 1 | 0.171 | 0 | 250 | 166.198407 | 50.008985 | 0.053009 |
5 rows × 42 columns
Next, we need to load the labels for the dataset. We import the labels.csv file using pandas and print the first five entries. The file column shows the relative path to the .dcm file and the label column indicates whether the chest x-ray has a pneumothorax or not.
df = pd.read_csv(pneumothorax_source/f"labels.csv")
df.head()| file | label | |
|---|---|---|
| 0 | train/No Pneumothorax/000000.dcm | No Pneumothorax |
| 1 | train/Pneumothorax/000001.dcm | Pneumothorax |
| 2 | train/No Pneumothorax/000002.dcm | No Pneumothorax |
| 3 | train/Pneumothorax/000003.dcm | Pneumothorax |
| 4 | train/Pneumothorax/000004.dcm | Pneumothorax |
Now, we use the DataBlock class to prepare the DICOM data for training.
As we are dealing with DICOM images, we need to use PILDicom as the ImageBlock category. This is so the DataBlock will know how to open the DICOM images. As this is a binary classification task we will use CategoryBlock
pneumothorax = DataBlock(blocks=(ImageBlock(cls=PILDicom), CategoryBlock),
get_x=lambda x:pneumothorax_source/f"{x[0]}",
get_y=lambda x:x[1],
batch_tfms=[*aug_transforms(size=224),Normalize.from_stats(*imagenet_stats)])
dls = pneumothorax.dataloaders(df.values, num_workers=0)Additionally, we plot a first batch with the specified transformations:
dls = pneumothorax.dataloaders(df.values)
dls.show_batch(max_n=16)Due to IPython and Windows limitation, python multiprocessing isn't available now.
So `number_workers` is changed to 0 to avoid getting stuckTraining
We can then use the vision_learner function and initiate the training.
learn = vision_learner(dls, resnet34, metrics=accuracy)Note that if you do not select a loss or optimizer function, fastai will try to choose the best selection for the task. You can check the loss function by calling loss_func
learn.loss_funcFlattenedLoss of CrossEntropyLoss()And you can do the same for the optimizer by calling opt_func
learn.opt_func<function fastai.optimizer.Adam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-05, wd=0.01, decouple_wd=True)>Use lr_find to try to find the best learning rate
learn.lr_find()SuggestedLRs(lr_min=0.005754399299621582, lr_steep=0.0063095735386013985)learn.fit_one_cycle(1)| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 1.191782 | 2.123666 | 0.320000 | 00:37 |
learn.predict(pneumothorax_source/f"train/Pneumothorax/000004.dcm")When predicting on an image learn.predict returns a tuple (class, class tensor and [probabilities of each class]).In this dataset there are only 2 classes No Pneumothorax and Pneumothorax hence the reason why each probability has 2 values, the first value is the probability whether the image belongs to class 0 or No Pneumothorax and the second value is the probability whether the image belongs to class 1 or Pneumothorax
tta = learn.tta(use_max=True)learn.show_results(max_n=16)interp = Interpretation.from_learner(learn)interp.plot_top_losses(2)Result Evaluation
Medical models are predominantly high impact so it is important to know how good a model is at detecting a certain condition.
This model has an accuracy of 56%. Accuracy can be defined as the number of correctly predicted data points out of all the data points. However in this context we can define accuracy as the probability that the model is correct and the patient has the condition PLUS the probability that the model is correct and the patient does not have the condition
There are some other key terms that need to be used when evaluating medical models:
False Positive & False Negative
False Positive is an error in which a test result improperly indicates presence of a condition, such as a disease (the result is positive), when in reality it is not present
False Negative is an error in which a test result improperly indicates no presence of a condition (the result is negative), when in reality it is present
Sensitivity & Specificity
- Sensitivity or True Positive Rate is where the model classifies a patient has the disease given the patient actually does have the disease. Sensitivity quantifies the avoidance of false negatives
Example: A new test was tested on 10,000 patients, if the new test has a sensitivity of 90% the test will correctly detect 9,000 (True Positive) patients but will miss 1000 (False Negative) patients that have the condition but were tested as not having the condition
- Specificity or True Negative Rate is where the model classifies a patient as not having the disease given the patient actually does not have the disease. Specificity quantifies the avoidance of false positives
Understanding and using sensitivity, specificity and predictive values is a great paper if you are interested in learning more about understanding sensitivity, specificity and predictive values.
PPV and NPV
Most medical testing is evaluated via PPV (Positive Predictive Value) or NPV (Negative Predictive Value).
PPV - if the model predicts a patient has a condition what is the probability that the patient actually has the condition
NPV - if the model predicts a patient does not have a condition what is the probability that the patient actually does not have the condition
The ideal value of the PPV, with a perfect test, is 1 (100%), and the worst possible value would be zero
The ideal value of the NPV, with a perfect test, is 1 (100%), and the worst possible value would be zero
Confusion Matrix
The confusion matrix is plotted against the valid dataset
interp = ClassificationInterpretation.from_learner(learn)
losses,idxs = interp.top_losses()
len(dls.valid_ds)==len(losses)==len(idxs)
interp.plot_confusion_matrix(figsize=(7,7))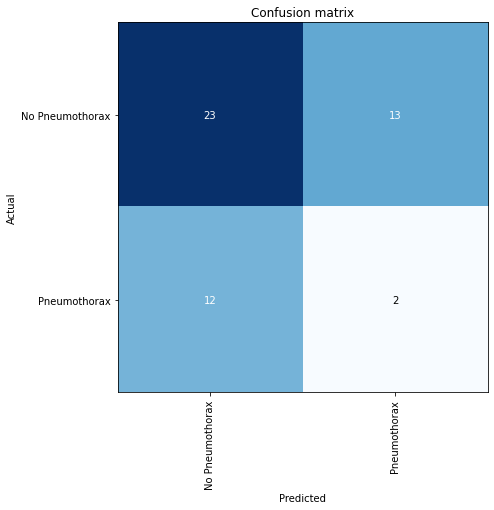
You can also reproduce the results interpreted from plot_confusion_matrix like so:
upp, low = interp.confusion_matrix()
tn, fp = upp[0], upp[1]
fn, tp = low[0], low[1]
print(tn, fp, fn, tp)23 13 12 2Note that Sensitivity = True Positive/(True Positive + False Negative)
sensitivity = tp/(tp + fn)
sensitivity0.14285714285714285In this case the model has a sensitivity of 40% and hence is only capable of correctly detecting 40% True Positives (i.e. who have Pneumothorax) but will miss 60% of False Negatives (patients that actually have Pneumothorax but were told they did not! Not a good situation to be in).
This is also know as a Type II error
Specificity = True Negative/(False Positive + True Negative)
specificity = tn/(fp + tn)
specificity0.6388888888888888The model has a specificity of 63% and hence can correctly detect 63% of the time that a patient does not have Pneumothorax but will incorrectly classify that 37% of the patients have Pneumothorax (False Postive) but actually do not.
This is also known as a Type I error
Positive Predictive Value (PPV)
ppv = tp/(tp+fp)
ppv0.13333333333333333In this case the model performs poorly in correctly predicting patients with Pneumothorax
Negative Predictive Value (NPV)
npv = tn/(tn+fn)
npv0.6571428571428571This model is better at predicting patients with No Pneumothorax
Calculating Accuracy
The accuracy of this model as mentioned before was 56% but how was this calculated? We can consider accuracy as:
accuracy = sensitivity x prevalence + specificity * (1 - prevalence)
Where prevalence is a statistical concept referring to the number of cases of a disease that are present in a particular population at a given time. The prevalence in this case is how many patients in the valid dataset have the condition compared to the total number.
To view the files in the valid dataset you call dls.valid_ds.cat
val = dls.valid_ds.cat
#val[0]There are 15 Pneumothorax images in the valid set (which has a total of 50 images and can be checked by using len(dls.valid_ds)) so the prevalence here is 15/50 = 0.3
prevalence = 15/50
prevalence0.3accuracy = (sensitivity * prevalence) + (specificity * (1 - prevalence))
accuracy0.490079365079365Citations:
[1] Filice R et al. Crowdsourcing pneumothorax annotations using machine learning annotations on the NIH chest X-ray dataset. J Digit Imaging (2019). https://doi.org/10.1007/s10278-019-00299-9