img = PILImage(PILImage.create(TEST_IMAGE).resize((600,400)))Vision augmentation
RandTransform-
RandTransform
def RandTransform(
p:float=1.0, # Probability of applying Transform
nm:str=None, before_call:Callable=None, # Optional batchwise preprocessing function
kwargs:VAR_KEYWORD
):
A transform that before_call its state at each __call__
As for all Transform you can pass encodes and decodes at init or subclass and implement them. You can do the same for the before_call method that is called at each __call__. Note that to have a consistent state for inputs and targets, a RandTransform must be applied at the tuple level.
By default the before_call behavior is to execute the transform with probability p (if subclassing and wanting to tweak that behavior, the attribute self.do, if it exists, is looked for to decide if the transform is executed or not).
A RandTransform is only applied to the training set by default, so you have to pass split_idx=0 if you are calling it directly and not through a Datasets. That behavior can be changed by setting the attr split_idx of the transform to None.
RandTransform.before_call<function __main__.RandTransform.before_call(self, b, split_idx: 'int')>RandTransform.before_call
def before_call(
b, split_idx:int, # Index of the train/valid dataset
):
This function can be overridden. Set self.do based on self.p
def _add1(x): return x+1
dumb_tfm = RandTransform(enc=_add1, p=0.5)
start,d1,d2 = 2,False,False
for _ in range(40):
t = dumb_tfm(start, split_idx=0)
if dumb_tfm.do: test_eq(t, start+1); d1=True
else: test_eq(t, start) ; d2=True
assert d1 and d2
dumb_tfm_add1 -- {'p': 0.5}
(enc:1,dec:0)Item transforms
FlipItem
def FlipItem(
p:float=0.5
):
Randomly flip with probability p
Calls @patch’d flip_lr behaviors for Image, TensorImage, TensorPoint, and TensorBBox
tflip = FlipItem(p=1.)
test_eq(tflip(bbox,split_idx=0), tensor([[1.,0., 0.,1]]) -1)DihedralItem
def DihedralItem(
p:float=1.0, # Probability of applying Transform
nm:str=None, before_call:Callable=None, # Optional batchwise preprocessing function
kwargs:VAR_KEYWORD
):
Randomly flip with probability p
Calls @patch’d dihedral behaviors for PILImage, TensorImage, TensorPoint, and TensorBBox
By default each of the 8 dihedral transformations (including noop) have the same probability of being picked when the transform is applied. You can customize this behavior by passing your own draw function. To force a specific flip, you can also pass an integer between 0 and 7.
_,axs = subplots(2, 4)
for ax in axs.flatten():
show_image(DihedralItem(p=1.)(img, split_idx=0), ctx=ax)
Resize with crop, pad or squish
PadMode
def PadMode(
args:VAR_POSITIONAL, kwargs:VAR_KEYWORD
):
All possible padding mode as attributes to get tab-completion and typo-proofing
CropPad
def CropPad(
size:int | tuple, # Size to crop or pad to, duplicated if one value is specified
pad_mode:PadMode='zeros', # A `PadMode`
enc:NoneType=None, dec:NoneType=None, split_idx:NoneType=None, order:NoneType=None
):
Center crop or pad an image to size
Calls @patch’d crop_pad behaviors for Image, TensorImage, TensorPoint, and TensorBBox
_,axs = plt.subplots(1,3,figsize=(12,4))
for ax,sz in zip(axs.flatten(), [300, 500, 700]):
show_image(img.crop_pad(sz), ctx=ax, title=f'Size {sz}');
print(img.crop_pad(sz).shape)(300, 300)
(500, 500)
(700, 700)
_,axs = plt.subplots(1,3,figsize=(12,4))
for ax,mode in zip(axs.flatten(), [PadMode.Zeros, PadMode.Border, PadMode.Reflection]):
show_image(img.crop_pad((600,700), pad_mode=mode), ctx=ax, title=mode);
RandomCrop
def RandomCrop(
size:int | tuple, # Size to crop to, duplicated if one value is specified
p:float=1.0, # Probability of applying Transform
nm:str=None, before_call:Callable=None, # Optional batchwise preprocessing function
):
Randomly crop an image to size
OldRandomCrop
def OldRandomCrop(
size:int | tuple, # Size to crop or pad to, duplicated if one value is specified
pad_mode:PadMode='zeros', # A `PadMode`
enc:NoneType=None, dec:NoneType=None, split_idx:NoneType=None, order:NoneType=None
):
Randomly crop an image to size
_,axs = plt.subplots(1,3,figsize=(12,4))
f = RandomCrop(200)
for ax in axs: show_image(f(img), ctx=ax);
On the validation set, we take a center crop.
_,axs = plt.subplots(1,3,figsize=(12,4))
for ax in axs: show_image(f(img, split_idx=1), ctx=ax);
ResizeMethod
def ResizeMethod(
args:VAR_POSITIONAL, kwargs:VAR_KEYWORD
):
All possible resize method as attributes to get tab-completion and typo-proofing
test_eq(ResizeMethod.Squish, 'squish')Resize
def Resize(
size:int | tuple, # Size to resize to, duplicated if one value is specified
method:ResizeMethod='crop', # A `ResizeMethod`
pad_mode:PadMode='reflection', # A `PadMode`
resamples:tuple=(<Resampling.BILINEAR: 2>, <Resampling.NEAREST: 0>), # Pillow `Image` resamples mode, resamples[1] for mask
p:float=1.0, # Probability of applying Transform
nm:str=None, before_call:Callable=None, # Optional batchwise preprocessing function
):
A transform that before_call its state at each __call__
size can be an integer (in which case images will be resized to a square) or a tuple. Depending on the method: - we squish any rectangle to size - we resize so that the shorter dimension is a match and use padding with pad_mode - we resize so that the larger dimension is match and crop (randomly on the training set, center crop for the validation set)
When doing the resize, we use resamples[0] for images and resamples[1] for segmentation masks.
_,axs = plt.subplots(1,3,figsize=(12,4))
for ax,method in zip(axs.flatten(), [ResizeMethod.Squish, ResizeMethod.Pad, ResizeMethod.Crop]):
rsz = Resize(256, method=method)
show_image(rsz(img, split_idx=0), ctx=ax, title=method);
On the validation set, the crop is always a center crop (on the dimension that’s cropped).
_,axs = plt.subplots(1,3,figsize=(12,4))
for ax,method in zip(axs.flatten(), [ResizeMethod.Squish, ResizeMethod.Pad, ResizeMethod.Crop]):
rsz = Resize(256, method=method)
show_image(rsz(img, split_idx=1), ctx=ax, title=method);
RandomResizedCrop
def RandomResizedCrop(
size:int | tuple, # Final size, duplicated if one value is specified,,
min_scale:float=0.08, # Minimum scale of the crop, in relation to image area
ratio:tuple=(0.75, 1.3333333333333333), # Range of width over height of the output
resamples:tuple=(<Resampling.BILINEAR: 2>, <Resampling.NEAREST: 0>), # Pillow `Image` resample mode, resamples[1] for mask
val_xtra:float=0.14, # The ratio of size at the edge cropped out in the validation set
max_scale:float=1.0, # Maximum scale of the crop, in relation to image area
p:float=1.0, # Probability of applying Transform
nm:str=None, before_call:Callable=None, # Optional batchwise preprocessing function
):
Picks a random scaled crop of an image and resize it to size
The crop picked as a random scale in range (min_scale,max_scale) and ratio in the range passed, then the resize is done with resamples[0] for images and resamples[1] for segmentation masks. On the validation set, we center crop the image if it’s ratio isn’t in the range (to the minmum or maximum value) then resize.
crop = RandomResizedCrop(256)
_,axs = plt.subplots(3,3,figsize=(9,9))
for ax in axs.flatten():
cropped = crop(img)
show_image(cropped, ctx=ax);
test_eq(cropped.shape, [256,256])Squish is used on the validation set, removing val_xtra proportion of each side first.
_,axs = subplots(1,3)
for ax in axs.flatten(): show_image(crop(img, split_idx=1), ctx=ax);
By setting max_scale to lower values, one can enforce small crops.
small_crop = RandomResizedCrop(256, min_scale=0.05, max_scale=0.15)
_,axs = plt.subplots(3,3,figsize=(9,9))
for ax in axs.flatten():
cropped = small_crop(img)
show_image(cropped, ctx=ax);
RatioResize
def RatioResize(
max_sz:int, # Biggest dimension of the resized image
resamples:tuple=(<Resampling.BILINEAR: 2>, <Resampling.NEAREST: 0>), # Pillow `Image` resample mode, resamples[1] for mask
kwargs:VAR_KEYWORD
):
Resizes the biggest dimension of an image to max_sz maintaining the aspect ratio
RatioResize(256)(img)
Affine and coord tfm on the GPU
timg = TensorImage(array(img)).permute(2,0,1).float()/255.
def _batch_ex(bs): return TensorImage(timg[None].expand(bs, *timg.shape).clone())Uses coordinates in coords to map coordinates in x to new locations for transformations such as flip. Preferably use TensorImage.affine_coord as this combines _grid_sample with F.affine_grid for easier usage. UseF.affine_grid to make it easier to generate the coords, as this tends to be large [H,W,2] where H and W are the height and width of your image x.
This is the image we start with, and are going to be using for the following examples.
img=torch.tensor([[[0,0,0],[1,0,0],[2,0,0]],
[[0,1,0],[1,1,0],[2,1,0]],
[[0,2,0],[1,2,0],[2,2,0]]]).permute(2,0,1)[None]/2.
show_images(img)
Here we _grid_sample, but do not change the original image. Notice how the coordinates in grid map to the coordiants in img.
grid=torch.tensor([[[[-1,-1],[0,-1],[1,-1]],
[[-1,0],[0,0],[1,0]],
[[-1,1],[0,1],[1,1.]]]])
img=_grid_sample(img, grid,align_corners=True)
show_images(img)
Next we do a flip by manually editing the grid.
grid=torch.tensor([[[1.,-1],[0,-1],[-1,-1]],
[[1,0],[0,0],[-1,0]],
[[1,1],[0,1],[-1,1]]])
img=_grid_sample(img, grid[None],align_corners=True)
show_images(img)
Next we shift the image up by one. By default _grid_sample uses reflection padding.
grid=torch.tensor([[[[-1,0],[0,0],[1,0]],
[[-1,1],[0,1],[1,1]],
[[-1,2],[0,2],[1,2.]]]])
img=_grid_sample(img, grid,align_corners=True)
show_images(img)
affine_coord allows us to much more easily work with images, by allowing us to specify much smaller mat, by comparison to grids, which require us to specify values for every pixel.
affine_grid
def affine_grid(
theta:Tensor, # Batch of affine transformation matrices
size:tuple, # Output size
align_corners:bool=None, # PyTorch `F.grid_sample` align_corners
):
Generates TensorFlowField from a transformation affine matrices theta
AffineCoordTfm
def AffineCoordTfm(
aff_fs:Callable | MutableSequence=None, # Affine transformations function for a batch
coord_fs:Callable | MutableSequence=None, # Coordinate transformations function for a batch
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
mode_mask:str='nearest', # Resample mode for mask
align_corners:NoneType=None, # PyTorch `F.grid_sample` align_corners
kwargs:VAR_KEYWORD
):
Combine and apply affine and coord transforms
Calls @patch’d affine_coord behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
Multiplies all the matrices returned by aff_fs before doing the corresponding affine transformation on a basic grid corresponding to size, then applies all coord_fs on the resulting flow of coordinates before finally doing an interpolation with mode and pad_mode.
Here are examples of how to use affine_coord on images. Including the identity or original image, a flip, and moving the image to the left.
imgs=_batch_ex(3)
identity=torch.tensor([[1,0,0],[0,1,0.]])
flip=torch.tensor([[-1,0,0],[0,1,0.]])
translation=torch.tensor([[1,0,1.],[0,1,0]])
mats=torch.stack((identity,flip,translation))
show_images(imgs.affine_coord(mats,pad_mode=PadMode.Zeros)) #Zeros easiest to see
Now you may be asking, “What is this mat”? Well lets take a quick look at the identify below.
imgs=_batch_ex(1)
identity=torch.tensor([[1,0,0],[0,1,0.]])
eye=identity[:,0:2]
bi=identity[:,2:3]
eye,bi(tensor([[1., 0.],
[0., 1.]]),
tensor([[0.],
[0.]]))Notice the tensor ‘eye’ is an identity matrix. If we multiply this by a single coordinate in our original image x,y we will simply the same values returned for x and y. bi is added after this multiplication. For example, lets flip the image so the left top corner is in the right top corner:
t=torch.tensor([[-1,0,0],[0,1,0.]])
eye=t[:,0:2]
bi=t[:,2:3]
xy=torch.tensor([-1.,-1]) #upper left corner
torch.sum(xy*eye,dim=1)+bi[0] #now the upper right cornertensor([ 1., -1.])AffineCoordTfm.compose
def compose(
tfm
):
Compose self with another AffineCoordTfm to only do the interpolation step once
RandomResizedCropGPU
def RandomResizedCropGPU(
size, # Final size, duplicated if one value is specified
min_scale:float=0.08, # Minimum scale of the crop, in relation to image area
ratio:tuple=(0.75, 1.3333333333333333), # Range of width over height of the output
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
valid_scale:float=1.0, # Scale of the crop for the validation set, in relation to image area
max_scale:float=1.0, # Maximum scale of the crop, in relation to image area
mode_mask:str='nearest', # Interpolation mode for [`TensorMask`](https://docs.fast.ai/torch_core.html#tensormask)
kwargs:VAR_KEYWORD
):
Picks a random scaled crop of an image and resize it to size
t = _batch_ex(8)
rrc = RandomResizedCropGPU(224, p=1.)
y = rrc(t)
_,axs = plt.subplots(2,4, figsize=(12,6))
for ax in axs.flatten():
show_image(y[i], ctx=ax)
RandomResizedCropGPU uses the same region for all images in the batch.
GPU helpers
This section contain helpers for working with augmentations on GPUs that is used throughout the code.
mask_tensor
def mask_tensor(
x:Tensor, # Input `Tensor`
p:float=0.5, # Probability of not applying mask
neutral:float=0.0, # Mask value
batch:bool=False, # Apply identical mask to entire batch
):
Mask elements of x with neutral with probability 1-p
Lets look at some examples of how mask_tensor might be used, we are using clone() because this operation overwrites the input. For this example lets try using degrees for rotating an image.
with no_random():
x=torch.tensor([60,-30,90,-210,270,-180,120,-240,150])
print('p=0.5: ',mask_tensor(x.clone()))
print('p=1.0: ',mask_tensor(x.clone(),p=1.))
print('p=0.0: ',mask_tensor(x.clone(),p=0.))p=0.5: tensor([ 60, -30, 90, -210, 0, -180, 0, 0, 150])
p=1.0: tensor([ 60, -30, 90, -210, 270, -180, 120, -240, 150])
p=0.0: tensor([0, 0, 0, 0, 0, 0, 0, 0, 0])Notice how p controls how likely a value is expected to be replaced with 0, or be unchanged since a 0 degree rotation would just be the original image. batch acts on the entire batch instead of single elements of the batch. Now lets consider a different example, of working with brightness. Note: with brightness 0 is a completely black image.
x=torch.tensor([0.6,0.4,0.3,0.7,0.4])
print('p=0.: ',mask_tensor(x.clone(),p=0))
print('p=0.,neutral=0.5: ',mask_tensor(x.clone(),p=0,neutral=0.5))p=0.: tensor([0., 0., 0., 0., 0.])
p=0.,neutral=0.5: tensor([0.5000, 0.5000, 0.5000, 0.5000, 0.5000])Here is would be very bad if we had a completely black image, as that is not an unchanged image. Instead we set neutral to 0.5 which is the value for an unchanged image for brightness.
_draw_mask is used to support the api of many following transformations to create mask_tensors. (p, neutral, batch) are passed down to mask_tensor. def_draw is the default draw function, and what should happen if no custom user setting is provided. draw is user defined behavior and can be a function, list of floats, or a float. draw and def_draw must return a tensor.
Here we use random integers from 1 to 8 for our def_draw, this example is very similar to Dihedral.
x = torch.zeros(10,2,3)
def def_draw(x):
x=torch.randint(1,8, (x.size(0),))
return x
with no_random(): print(torch.randint(1,8, (x.size(0),)))
with no_random(): print(_draw_mask(x, def_draw))tensor([2, 3, 5, 6, 5, 4, 6, 6, 1, 1])
TensorBase([2, 0, 0, 6, 5, 4, 6, 0, 0, 1])Next, there are three ways to define draw, as a constant, as a list, and as a function. All of these override def_draw, so that it has no effect on the final result.
with no_random():
print('const: ',_draw_mask(x, def_draw, draw=1))
print('list : ', _draw_mask(x, def_draw, draw=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]))
print('list : ',_draw_mask(x[0:2], def_draw, draw=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14]))
print('funct: ',_draw_mask(x, def_draw, draw=lambda x: torch.arange(1,x.size(0)+1)))
try:
_draw_mask(x, def_draw, draw=[1,2])
except AssertionError as e:
print(type(e),'\n',e)const: TensorBase([1., 1., 1., 1., 0., 1., 0., 0., 1., 1.])
list : TensorBase([ 1., 2., 0., 0., 5., 0., 7., 0., 0., 10.])
list : TensorBase([1., 0.])
funct: TensorBase([ 1, 2, 3, 4, 0, 6, 7, 8, 9, 10])
<class 'AssertionError'>
Note, when using a list it can be larger than the batch size, but it cannot be smaller than the batch size. Otherwise there would not be enough augmentations for elements of the batch.
x = torch.zeros(5,2,3)
def_draw = lambda x: torch.randint(0,8, (x.size(0),))
t = _draw_mask(x, def_draw)
assert (0. <= t).all() and (t <= 7).all()
t = _draw_mask(x, def_draw, 1)
assert (0. <= t).all() and (t <= 1).all()
test_eq(_draw_mask(x, def_draw, 1, p=1), tensor([1.,1,1,1,1]))
test_eq(_draw_mask(x, def_draw, [0,1,2,3,4], p=1), tensor([0.,1,2,3,4]))
test_eq(_draw_mask(x[0:3], def_draw, [0,1,2,3,4], p=1), tensor([0.,1,2]))
for i in range(5):
t = _draw_mask(x, def_draw, 1,batch=True)
assert (t==torch.zeros(5)).all() or (t==torch.ones(5)).all()Flip/Dihedral GPU Helpers
affine_mat is used to transform the length-6 vestor into a [bs,3,3] tensor. This is used to allow us to combine affine transforms.
affine_mat
def affine_mat(
ms:VAR_POSITIONAL
):
Restructure length-6 vector ms into an affine matrix with 0,0,1 in the last line
Here is an example of how flipping an image would look using affine_mat.
flips=torch.tensor([-1,1,-1])
ones=t1(flips)
zeroes=t0(flips)
affines=affine_mat(flips,zeroes,zeroes,zeroes,ones,zeroes)
print(affines)tensor([[[-1, 0, 0],
[ 0, 1, 0],
[ 0, 0, 1]],
[[ 1, 0, 0],
[ 0, 1, 0],
[ 0, 0, 1]],
[[-1, 0, 0],
[ 0, 1, 0],
[ 0, 0, 1]]])This is done so that we can combine multiple affine transformations without doing the math on the entire image. We need the matrices to be the same size, so we can do a matric multiple in order to combines affine transformations. While this is usually done on an entire batch, here is what it would look like to have multiple flip transformations for a single image. Since we flip twice we end up with an affine matrix that would simply return our original image.
If you would like more information on how this works, see affine_coord.
x = torch.eye(3,dtype=torch.int64)
for affine in affines:
x @= affine
print(x)tensor([[-1, 0, 0],
[ 0, 1, 0],
[ 0, 0, 1]])
tensor([[-1, 0, 0],
[ 0, 1, 0],
[ 0, 0, 1]])
tensor([[1, 0, 0],
[0, 1, 0],
[0, 0, 1]])flip_mat will generate a [bs,3,3] tensor representing our flips for a batch with probability p. draw can be used to define a function, constant, or list that defines what flips to use. If draw is a list, the length must be greater than or equal to the batch size. For draw 0 is the original image, or 1 is a flipped image. batch will mean that the entire batch will be flipped or not.
flip_mat
def flip_mat(
x:Tensor, # The input Tensor
p:float=0.5, # Probability of appying transformation
draw:int | MutableSequence | Callable=None, # Custom flips instead of random
batch:bool=False, # Apply identical flip to entire batch
):
Return a random flip matrix
Below are some examples of how to use draw as a constant, list and function.
with no_random():
x=torch.randn(2,4,3)
print('const: ',flip_mat(x, draw=1))
print('list : ', flip_mat(x, draw=[1, 0]))
print('list : ',flip_mat(x[0:2], draw=[1, 0, 1, 0, 1]))
print('funct: ',flip_mat(x, draw=lambda x: torch.ones(x.size(0))))
test_fail(lambda: flip_mat(x, draw=[1]))const: TensorBase([[[-1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]],
[[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]]])
list : TensorBase([[[-1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]],
[[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]]])
list : TensorBase([[[-1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]],
[[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]]])
funct: TensorBase([[[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]],
[[-1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]]])x = flip_mat(torch.randn(100,4,3))
test_eq(set(x[:,0,0].numpy()), {-1,1}) #might fail with probability 2*2**(-100) (picked only 1s or -1s)Flip images,masks,points and bounding boxes horizontally. p is the probability of a flip being applied. draw can be used to define custom flip behavior.
Flip
def Flip(
p:float=0.5, # Probability of applying flip
draw:int | MutableSequence | Callable=None, # Custom flips instead of random
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
batch:bool=False, # Apply identical flip to entire batch
):
Randomly flip a batch of images with a probability p
Calls @patch’d flip_batch behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
Here are some examples of using flip. Notice that a constant draw=1, is effectively the same as the default settings. Also notice the fine-tune control we can get in the third example, by setting p=1. and defining a custom draw.
with no_random(32):
imgs = _batch_ex(5)
deflt = Flip()
const = Flip(p=1.,draw=1) #same as default
listy = Flip(p=1.,draw=[1,0,1,0,1]) #completely manual!!!
funct = Flip(draw=lambda x: torch.ones(x.size(0))) #same as default
show_images( deflt(imgs) ,suptitle='Default Flip')
show_images( const(imgs) ,suptitle='Constant Flip',titles=[f'Flipped' for i in['','','','','']]) #same above
show_images( listy(imgs) ,suptitle='Listy Flip',titles=[f'{i}Flipped' for i in ['','Not ','','Not ','']])
show_images( funct(imgs) ,suptitle='Flip By Function') #same as default



flip = Flip(p=1.)
t = _pnt2tensor([[1,0], [2,1]], (3,3))
y = flip(TensorImage(t[None,None]), split_idx=0)
test_eq(y, _pnt2tensor([[1,0], [0,1]], (3,3))[None,None])
pnts = TensorPoint((tensor([[1.,0.], [2,1]]) -1)[None])
test_eq(flip(pnts, split_idx=0), tensor([[[1.,0.], [0,1]]]) -1)
bbox = TensorBBox(((tensor([[1.,0., 2.,1]]) -1)[None]))
test_eq(flip(bbox, split_idx=0), tensor([[[0.,0., 1.,1.]]]) -1)DeterministicDraw
def DeterministicDraw(
vals
):
Initialize self. See help(type(self)) for accurate signature.
t = _batch_ex(8)
draw = DeterministicDraw(list(range(8)))
for i in range(15): test_eq(draw(t), torch.zeros(8)+(i%8))DeterministicFlip
def DeterministicFlip(
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
kwargs:VAR_KEYWORD
):
Flip the batch every other call
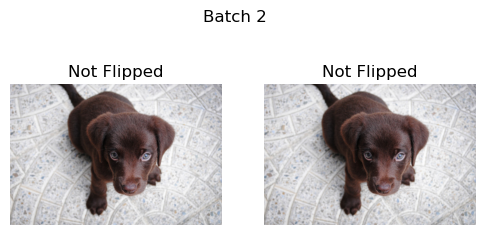
Next we loop through multiple batches of the example images. DeterministicFlip will first not flip the images, and then on the next batch it will flip the images.
b = _batch_ex(2)
dih = DeterministicFlip()
for i,flipped in enumerate(['Not Flipped','Flipped']*2):
show_images(dih(b),suptitle=f'Batch {i}',titles=[flipped]*2)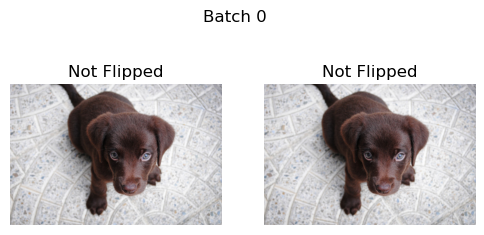


Since we are working with squares and rectangles, we can think of dihedral flips as flips across the horizontal, vertical, and diagonal and their combinations. Remember though that rectangles are not symmetrical across their diagonal, so this will effectively cropping parts of rectangles.
dihedral_mat
def dihedral_mat(
x:Tensor, # Input `Tensor`
p:float=0.5, # Probability of staying unchanged
draw:int | MutableSequence | Callable=None, # Custom dihedrals instead of random
batch:bool=False, # Apply identical dihedral to entire batch
):
Return a random dihedral matrix
Dihedral
def Dihedral(
p:float=0.5, # Probability of applying dihedral
draw:int | MutableSequence | Callable=None, # Custom dihedrals instead of random
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
batch:bool=False, # Apply identical dihedral to entire batch
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
):
Apply a random dihedral transformation to a batch of images with a probability p
Calls @patch’d dihedral_batch behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
draw can be specified if you want to customize which flip is picked when the transform is applied (default is a random number between 0 and 7). It can be an integer between 0 and 7, a list of such integers (which then should have a length equal to or greater than the size of the batch) or a callable that returns a long tensor between 0 and 7.
with no_random():
imgs = _batch_ex(5)
deflt = Dihedral()
const = Dihedral(p=1.,draw=1) #same as flip_batch
listy = Dihedral(p=1.,draw=[0,1,2,3,4]) #completely manual!!!
funct = Dihedral(draw=lambda x: torch.randint(0,8,(x.size(0),))) #same as default
show_images( deflt(imgs) ,suptitle='Default Flips',titles=[i for i in range(imgs.size(0))])
show_images( const(imgs) ,suptitle='Constant Horizontal Flip',titles=[f'Flip 1' for i in [0,1,1,1,1]])
show_images( listy(imgs) ,suptitle='Manual Listy Flips',titles=[f'Flip {i}' for i in [0,1,2,3,4]]) #manually specified, not random!
show_images( funct(imgs) ,suptitle='Default Functional Flips',titles=[i for i in range(imgs.size(0))]) #same as default



DeterministicDihedral
def DeterministicDihedral(
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
align_corners:NoneType=None, # PyTorch `F.grid_sample` align_corners
):
Apply a random dihedral transformation to a batch of images with a probability p
DeterministicDihedral guarantees that the first call will not be flipped, then the following call will be flip in a deterministic order. After all 7 possible dihedral flips the pattern will reset to the unflipped version. If we were to do this on a batch size of one it would look like this:
t = _batch_ex(10)
dih = DeterministicDihedral()
_,axs = plt.subplots(2,5, figsize=(14,6))
for i,ax in enumerate(axs.flatten()):
y = dih(t)
show_image(y[0], ctx=ax, title=f'Batch {i}')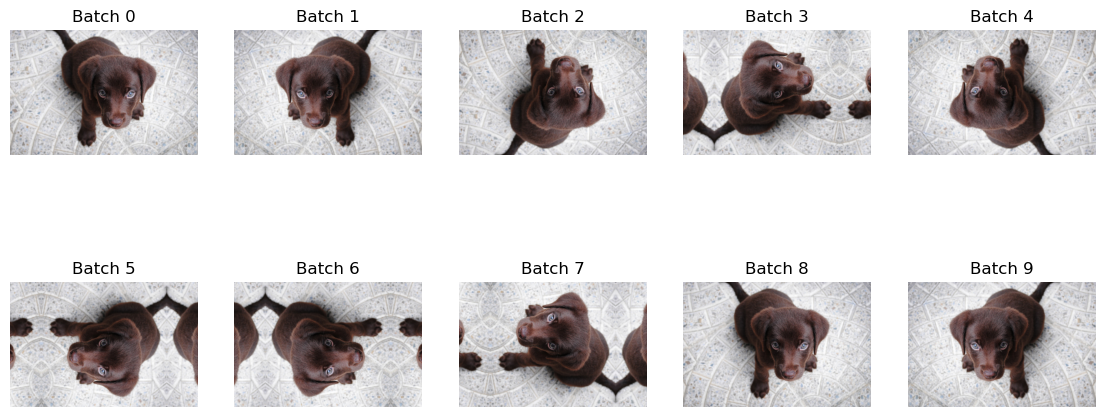
rotate_mat
def rotate_mat(
x:Tensor, # Input `Tensor`
max_deg:int=10, # Maximum degree of rotation
p:float=0.5, # Probability of applying rotate
draw:int | MutableSequence | Callable=None, # Custom rotates instead of random
batch:bool=False, # Apply identical rotate to entire batch
):
Return a random rotation matrix with max_deg and p
Rotate
def Rotate(
max_deg:int=10, # Maximum degree of rotation
p:float=0.5, # Probability of applying rotate
draw:int | MutableSequence | Callable=None, # Custom rotates instead of random
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
batch:bool=False, # Apply identical rotate to entire batch
):
Apply a random rotation of at most max_deg with probability p to a batch of images
Calls @patch’d rotate behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
draw can be specified if you want to customize which angle is picked when the transform is applied (default is a random float between -max_deg and max_deg). It can be a float, a list of floats (which then should have a length equal to or greater than the size of the batch) or a callable that returns a float tensor.
Rotate by default can only rotate 10 degrees, which makes the changes harder to see. This is usually combined with either flip or dihedral, which make much larger changes by default. A rotate of 180 degrees is the same as a vertical flip for example.
with no_random():
thetas = [-30,-15,0,15,30]
imgs = _batch_ex(5)
deflt = Rotate()
const = Rotate(p=1.,draw=180) #same as a vertical flip
listy = Rotate(p=1.,draw=[-30,-15,0,15,30]) #completely manual!!!
funct = Rotate(draw=lambda x: x.new_empty(x.size(0)).uniform_(-10, 10)) #same as default
show_images( deflt(imgs) ,suptitle='Default Rotate, notice the small rotation',titles=[i for i in range(imgs.size(0))])
show_images( const(imgs) ,suptitle='Constant 180 Rotate',titles=[f'180 Degrees' for i in range(imgs.size(0))])
#manually specified, not random!
show_images( listy(imgs) ,suptitle='Manual List Rotate',titles=[f'{i} Degrees' for i in [-30,-15,0,15,30]])
#same as default
show_images( funct(imgs) ,suptitle='Default Functional Rotate',titles=[i for i in range(imgs.size(0))])



zoom_mat
def zoom_mat(
x:Tensor, # Input `Tensor`
min_zoom:float=1.0, # Minimum zoom
max_zoom:float=1.1, # Maximum zoom
p:float=0.5, # Probability of applying zoom
draw:float | MutableSequence | Callable=None, # User defined scale of the zoom
draw_x:float | MutableSequence | Callable=None, # User defined center of the zoom in x
draw_y:float | MutableSequence | Callable=None, # User defined center of the zoom in y
batch:bool=False, # Apply identical zoom to entire batch
):
Return a random zoom matrix with max_zoom and p
Zoom
def Zoom(
min_zoom:float=1.0, # Minimum zoom
max_zoom:float=1.1, # Maximum zoom
p:float=0.5, # Probability of applying zoom
draw:float | MutableSequence | Callable=None, # User defined scale of the zoom
draw_x:float | MutableSequence | Callable=None, # User defined center of the zoom in x
draw_y:float | MutableSequence | Callable=None, # User defined center of the zoom in y
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
batch:bool=False, # Apply identical zoom to entire batch
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
):
Apply a random zoom of at most max_zoom with probability p to a batch of images
Calls @patch’d zoom behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
draw, draw_x and draw_y can be specified if you want to customize which scale and center are picked when the transform is applied (default is a random float between 1 and max_zoom for the first, between 0 and 1 for the last two). Each can be a float, a list of floats (which then should have a length equal to or greater than the size of the batch) or a callable that returns a float tensor.
draw_x and draw_y are expected to be the position of the center in pct, 0 meaning the most left/top possible and 1 meaning the most right/bottom possible.
Note: By default Zooms are rather small.
with no_random():
scales = [0.8, 1., 1.1, 1.25, 1.5]
imgs = _batch_ex(5)
deflt = Zoom()
const = Zoom(p=1., draw=1.5) #'Constant scale and different random centers'
listy = Zoom(p=1.,draw=scales,draw_x=0.5, draw_y=0.5) #completely manual scales, constant center
funct = Zoom(draw=lambda x: x.new_empty(x.size(0)).uniform_(1., 1.1)) #same as default
show_images( deflt(imgs) ,suptitle='Default Zoom, note the small zooming', titles=[i for i in range(imgs.size(0))])
show_images( const(imgs) ,suptitle='Constant Scale, Valiable Position', titles=[f'Scale 1.5x' for i in range(imgs.size(0))])
show_images( listy(imgs) ,suptitle='Manual Listy Scale, Centered', titles=[f'Scale {i}x' for i in scales])
show_images( funct(imgs) ,suptitle='Default Functional Zoom', titles=[i for i in range(imgs.size(0))]) #same as default



Warping
find_coeffs
def find_coeffs(
p1:Tensor, # Original points
p2:Tensor, # Target points
):
Find coefficients for warp tfm from p1 to p2
apply_perspective
def apply_perspective(
coords:Tensor, # Original coordinates
coeffs:Tensor, # Warping transformation matrice
):
Apply perspective tranform on coords with coeffs
Warp
def Warp(
magnitude:float=0.2, # The default warping magnitude
p:float=0.5, # Probability of applying warp
draw_x:float | MutableSequence | Callable=None, # User defined warping magnitude in x
draw_y:float | MutableSequence | Callable=None, # User defined warping magnitude in y
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
batch:bool=False, # Apply identical warp to entire batch
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
):
Apply perspective warping with magnitude and p on a batch of matrices
Calls @patch’d warp behaviors for TensorImage, TensorMask, TensorPoint, and TensorBBox
draw_x and draw_y can be specified if you want to customize the magnitudes that are picked when the transform is applied (default is a random float between -magnitude and magnitude. Each can be a float, a list of floats (which then should have a length equal to or greater than the size of the batch) or a callable that returns a float tensor.
scales = [-0.4, -0.2, 0., 0.2, 0.4]
imgs=_batch_ex(5)
vert_warp = Warp(p=1., draw_y=scales, draw_x=0.)
horz_warp = Warp(p=1., draw_x=scales, draw_y=0.)
show_images( vert_warp(imgs) ,suptitle='Vertical warping', titles=[f'magnitude {i}' for i in scales])
show_images( horz_warp(imgs) ,suptitle='Horizontal warping', titles=[f'magnitude {i}' for i in scales])

Lighting transforms
Lighting transforms are transforms that effect how light is represented in an image. These don’t change the location of the object like previous transforms, but instead simulate how light could change in a scene. The simclr paper evaluates these transforms against other transforms for their use case of self-supurved image classification, note they use “color” and “color distortion” to refer to a combination of these transforms.
TensorImage.lighting
def lighting(
x:TensorImage, func
):
Most lighting transforms work better in “logit space”, as we do not want to blowout the image by going over maximum or minimum brightness. Taking the sigmoid of the logit allows us to get back to “linear space.”
x=TensorImage(torch.tensor([.01* i for i in range(0,101)]))
f_lin= lambda x:(2*(x-0.5)+0.5).clamp(0,1) #blue line
f_log= lambda x:2*x #red line
plt.plot(x,f_lin(x),'b',x,x.lighting(f_log),'r');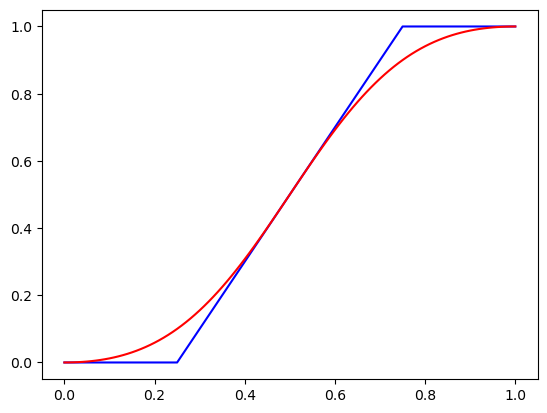
The above graph shows the results of doing a contrast transformation in both linear and logit space. Notice how the blue linear plot has to be clamped, and we have lost information on how large 0.0 is by comparision to 0.2. While in the red plot the values curve, so we keep this relative information.
First we create a general SpaceTfm. This allows us compose multiple transforms together, so that we only have to convert to a space once, before doing multiple transforms. The space_fn must convert from rgb to a space, apply a function, and then convert back to rgb. fs should be list-like, and contain a functions that will be composed together.
SpaceTfm
def SpaceTfm(
fs:Callable | MutableSequence, # Transformation functions applying in a space
space_fn:Callable, # Function converting rgb to a space and back to rgb after appying `fs`
kwargs:VAR_KEYWORD
):
Apply fs to the logits
LightingTfm is a SpaceTfm that uses TensorImage.lighting to convert to logit space. Use this to limit images loosing detail when they become very dark or bright.
LightingTfm
def LightingTfm(
fs:Callable | MutableSequence, # Transformation functions applying in logit space,
kwargs:VAR_KEYWORD
):
Apply fs to the logits
Brightness refers to the amount of light on a scene. This can be zero in which the image is completely black or one where the image is completely white. This may be especially useful if you expect your dataset to have over or under exposed images.
Brightness
def Brightness(
max_lighting:float=0.2, # Maximum scale of changing brightness
p:float=0.75, # Probability of appying transformation
draw:float | MutableSequence | Callable=None, # User defined behavior of batch transformation
batch:bool=False, # Apply identical brightness to entire batch
):
Apply fs to the logits
Calls @patch’d brightness behaviors for TensorImage
draw can be specified if you want to customize the magnitude that is picked when the transform is applied (default is a random float between -0.5*(1-max_lighting) and 0.5*(1+max_lighting). Each can be a float, a list of floats (which then should have a length equal to or greater than the size of the batch) or a callable that returns a float tensor.
scales = [0.1, 0.3, 0.5, 0.7, 0.9]
y = _batch_ex(5).brightness(draw=scales, p=1.)
fig,axs = plt.subplots(1,5, figsize=(15,3))
for i,ax in enumerate(axs.flatten()):
show_image(y[i], ctx=ax, title=f'scale {scales[i]}')
Contrast pushes pixels to either the maximum or minimum values. The minimum value for contrast is a solid gray image. As an example take a picture of a bright light source in a dark room. Your eyes should be able to see some detail in the room, but the photo taken should instead have much higher contrast, with all of the detail in the background missing to the darkness. This is one example of what this transform can help simulate.
Contrast
def Contrast(
max_lighting:float=0.2, # Maximum scale of changing contrast
p:float=0.75, # Probability of appying transformation
draw:float | MutableSequence | Callable=None, # User defined behavior of batch transformation
batch:bool=False
):
Apply change in contrast of max_lighting to batch of images with probability p.
Calls @patch’d contrast behaviors for TensorImage
draw can be specified if you want to customize the magnitude that is picked when the transform is applied (default is a random float taken with the log uniform distribution between (1-max_lighting) and 1/(1-max_lighting). Each can be a float, a list of floats (which then should have a length equal to or greater than the size of the batch) or a callable that returns a float tensor.
scales = [0.65, 0.8, 1., 1.25, 1.55]
y = _batch_ex(5).contrast(p=1., draw=scales)
fig,axs = plt.subplots(1,5, figsize=(15,3))
for i,ax in enumerate(axs.flatten()): show_image(y[i], ctx=ax, title=f'scale {scales[i]}')
grayscale
def grayscale(
x
):
Tensor to grayscale tensor. Uses the ITU-R 601-2 luma transform.
The above is just one way to convert to grayscale. We chose this one because it was fast. Notice that the sum of the weight of each channel is 1.
f'{sum([0.2989,0.5870,0.1140]):.3f}''1.000'Saturation
def Saturation(
max_lighting:float=0.2, # Maximum scale of changing brightness
p:float=0.75, # Probability of appying transformation
draw:float | MutableSequence | Callable=None, # User defined behavior of batch transformation
batch:bool=False, # Apply identical saturation to entire batch
):
Apply change in saturation of max_lighting to batch of images with probability p.
Calls @patch’d saturation behaviors for TensorImage
scales = [0., 0.5, 1., 1.5, 2.0]
y = _batch_ex(5).saturation(p=1., draw=scales)
fig,axs = plt.subplots(1,5, figsize=(15,3))
for i,ax in enumerate(axs.flatten()): show_image(y[i], ctx=ax, title=f'scale {scales[i]}')
Saturation controls the amount of color in the image, but not the lightness or darkness of an image. If has no effect on neutral colors such as whites,grays and blacks. At zero saturation you actually get a grayscale image. Pushing saturation past one causes more neutral colors to take on any underlying chromatic color.
rgb2hsv, and hsv2rgb are utilities for converting to and from hsv space. Hsv space stands for hue,saturation, and value space. This allows us to more easily perform certain transforms.
torch.max(tensor([1]).as_subclass(TensorBase), dim=0)torch.return_types.max(
values=TensorBase(1),
indices=TensorBase(0))rgb2hsv
def rgb2hsv(
img:Tensor, # Batch of images `Tensor`in RGB
):
Converts a RGB image to an HSV image. Note: Will not work on logit space images.
hsv2rgb
def hsv2rgb(
img:Tensor, # Batch of images `Tensor in HSV`
):
Converts a HSV image to an RGB image.
Very similar to lighting which is done in logit space, hsv transforms are done in hsv space. We can compose any transforms that are done in hsv space.
HSVTfm
def HSVTfm(
fs, kwargs:VAR_KEYWORD
):
Apply fs to the images in HSV space
Calls @patch’d hsv behaviors for TensorImage
fig,axs=plt.subplots(figsize=(20, 4),ncols=5)
axs[0].set_ylabel('Hue')
for ax in axs:
ax.set_xlabel('Saturation')
ax.set_yticklabels([])
ax.set_xticklabels([])
hsvs=torch.stack([torch.arange(0,2.1,0.01)[:,None].repeat(1,210),
torch.arange(0,1.05,0.005)[None].repeat(210,1),
torch.ones([210,210])])[None]
for ax,i in zip(axs,range(0,5)):
if i>0: hsvs[:,2].mul_(0.80)
ax.set_title('V='+'%.1f' %0.8**i)
ax.imshow(hsv2rgb(hsvs)[0].permute(1,2,0))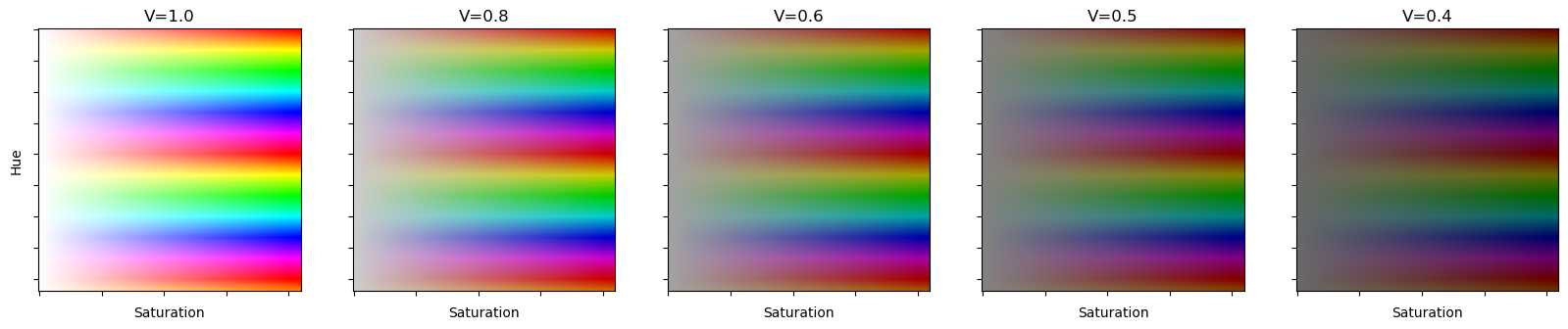
For the Hue transform we are using hsv space instead of logit space. HSV stands for hue,saturation and value. Hue in hsv space just cycles through colors of the rainbow. Notices how there is no maximum, because the colors just repeat.
Above are some examples of Hue(H) and Saturation(S) at various Values(V). One property of note in HSV space is that V controls the color you get at minimum saturation when in HSV space.
Hue
def Hue(
max_hue:float=0.1, # Maximum scale of changing Hue
p:float=0.75, # Probability of appying transformation
draw:float | MutableSequence | Callable=None, # User defined behavior of batch transformation
batch:bool=False, # Apply identical Hue to entire batch
):
Apply change in hue of max_hue to batch of images with probability p.
Calls @patch’d hue behaviors for TensorImage
scales = [0.5, 0.75, 1., 1.5, 1.75]
y = _batch_ex(len(scales)).hue(p=1., draw=scales)
fig,axs = plt.subplots(1,len(scales), figsize=(15,3))
for i,ax in enumerate(axs.flatten()): show_image(y[i], ctx=ax, title=f'scale {scales[i]}')
RandomErasing
Random Erasing Data Augmentation. This variant, designed by Ross Wightman, is applied to either a batch or single image tensor after it has been normalized.
cutout_gaussian
def cutout_gaussian(
x:Tensor, # Input image
areas:list, # List of areas to cutout. Order rl,rh,cl,ch
):
Replace all areas in x with N(0,1) noise
Since this should be applied after normalization, we’ll define a helper to apply a function inside normalization.
norm_apply_denorm
def norm_apply_denorm(
x:Tensor, # Input Image
f:Callable, # Function to apply
nrm:Callable, # Normalization transformation
):
Normalize x with nrm, then apply f, then denormalize
nrm = Normalize.from_stats(*imagenet_stats, cuda=False)f = partial(cutout_gaussian, areas=[(100,200,100,200),(200,300,200,300)])
show_image(norm_apply_denorm(timg, f, nrm)[0]);
RandomErasing
def RandomErasing(
p:float=0.5, # Probability of appying Random Erasing
sl:float=0.0, # Minimum proportion of erased area
sh:float=0.3, # Maximum proportion of erased area
min_aspect:float=0.3, # Minimum aspect ratio of erased area
max_count:int=1, # Maximum number of erasing blocks per image, area per box is scaled by count
):
Randomly selects a rectangle region in an image and randomizes its pixels.
tfm = RandomErasing(p=1., max_count=6)
_,axs = subplots(2,3, figsize=(12,6))
f = partial(tfm, split_idx=0)
for i,ax in enumerate(axs.flatten()): show_image(norm_apply_denorm(timg, f, nrm)[0], ctx=ax)
tfm = RandomErasing(p=1., max_count=6)
_,axs = subplots(2,3, figsize=(12,6))
f = partial(tfm, split_idx=0)
for i,ax in enumerate(axs.flatten()): show_image(norm_apply_denorm(timg, f, nrm)[0], ctx=ax)
tfm = RandomErasing(p=1., max_count=6)
_,axs = subplots(2,3, figsize=(12,6))
f = partial(tfm, split_idx=1)
for i,ax in enumerate(axs.flatten()): show_image(norm_apply_denorm(timg, f, nrm)[0], ctx=ax)
All together
setup_aug_tfms
def setup_aug_tfms(
tfms
):
Go through tfms and combines together affine/coord or lighting transforms
#Affine only
tfms = [Rotate(draw=10., p=1), Zoom(draw=1.1, draw_x=0.5, draw_y=0.5, p=1.)]
comp = setup_aug_tfms([Rotate(draw=10., p=1), Zoom(draw=1.1, draw_x=0.5, draw_y=0.5, p=1.)])
test_eq(len(comp), 1)
x = torch.randn(4,3,5,5)
test_close(comp[0]._get_affine_mat(x)[...,:2],tfms[0]._get_affine_mat(x)[...,:2] @ tfms[1]._get_affine_mat(x)[...,:2])
#We can't test that the ouput of comp or the composition of tfms on x is the same cause it's not (1 interpol vs 2 sp)#Affine + lighting
tfms = [Rotate(), Zoom(), Warp(), Brightness(), Flip(), Contrast()]
comp = setup_aug_tfms(tfms)aff_tfm,lig_tfm = comp
test_eq(len(aff_tfm.aff_fs+aff_tfm.coord_fs+comp[1].fs), 6)
test_eq(len(aff_tfm.aff_fs), 3)
test_eq(len(aff_tfm.coord_fs), 1)
test_eq(len(lig_tfm.fs), 2)aug_transforms
def aug_transforms(
mult:float=1.0, # Multiplication applying to `max_rotate`,`max_lighting`,`max_warp`
do_flip:bool=True, # Random flipping
flip_vert:bool=False, # Flip vertically
max_rotate:float=10.0, # Maximum degree of rotation
min_zoom:float=1.0, # Minimum zoom
max_zoom:float=1.1, # Maximum zoom
max_lighting:float=0.2, # Maximum scale of changing brightness
max_warp:float=0.2, # Maximum value of changing warp per
p_affine:float=0.75, # Probability of applying affine transformation
p_lighting:float=0.75, # Probability of changing brightnest and contrast
xtra_tfms:list=None, # Custom Transformations
size:int | tuple=None, # Output size, duplicated if one value is specified
mode:str='bilinear', # PyTorch `F.grid_sample` interpolation
pad_mode:str='reflection', # A `PadMode`
align_corners:bool=True, # PyTorch `F.grid_sample` align_corners
batch:bool=False, # Apply identical transformation to entire batch
min_scale:float=1.0, # Minimum scale of the crop, in relation to image area
):
Utility func to easily create a list of flip, rotate, zoom, warp, lighting transforms.
Random flip (or dihedral if flip_vert=True) with p=0.5 is added when do_flip=True. With p_affine we apply a random rotation of max_rotate degrees, a random zoom between min_zoom and max_zoom and a perspective warping of max_warp. With p_lighting we apply a change in brightness and contrast of max_lighting. Custom xtra_tfms can be added. size, mode and pad_mode will be used for the interpolation. max_rotate,max_lighting,max_warp are multiplied by mult so you can more easily increase or decrease augmentation with a single parameter.
tfms = aug_transforms(pad_mode='zeros', mult=2, min_scale=0.5)
y = _batch_ex(9)
for t in tfms: y = t(y, split_idx=0)
_,axs = plt.subplots(1,3, figsize=(12,3))
for i,ax in enumerate(axs.flatten()): show_image(y[i], ctx=ax)
tfms = aug_transforms(pad_mode='zeros', mult=2, batch=True)
y = _batch_ex(9)
for t in tfms: y = t(y, split_idx=0)
_,axs = plt.subplots(1,3, figsize=(12,3))
for i,ax in enumerate(axs.flatten()): show_image(y[i], ctx=ax)
Integration tests
Segmentation
camvid = untar_data(URLs.CAMVID_TINY)
fns = get_image_files(camvid/'images')
cam_fn = fns[0]
mask_fn = camvid/'labels'/f'{cam_fn.stem}_P{cam_fn.suffix}'
def _cam_lbl(fn): return mask_fncam_dsrc = Datasets([cam_fn]*10, [PILImage.create, [_cam_lbl, PILMask.create]])
cam_tdl = TfmdDL(cam_dsrc.train, after_item=ToTensor(),
after_batch=[IntToFloatTensor(), *aug_transforms()], bs=9)
cam_tdl.show_batch(max_n=9, vmin=1, vmax=30)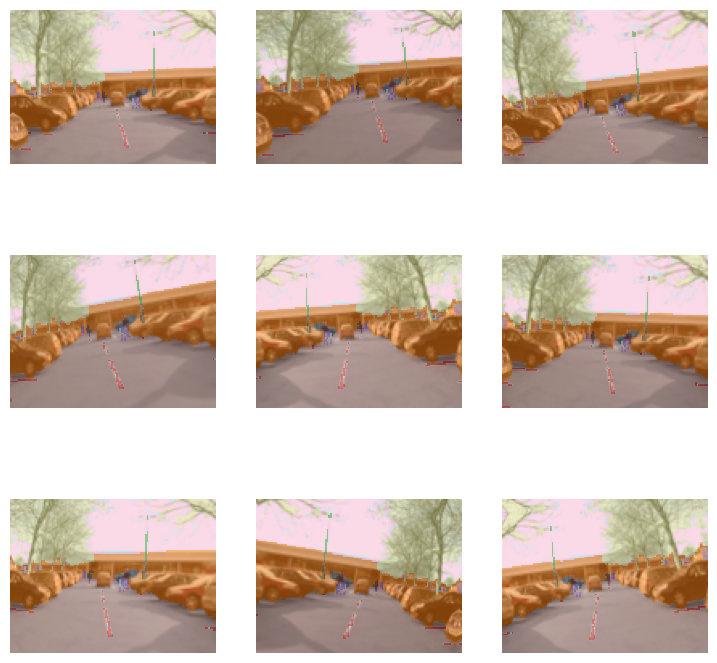
Point targets
mnist = untar_data(URLs.MNIST_TINY)
mnist_fn = 'images/mnist3.png'
pnts = np.array([[0,0], [0,35], [28,0], [28,35], [9, 17]])
def _pnt_lbl(fn)->None: return TensorPoint.create(pnts)pnt_dsrc = Datasets([mnist_fn]*10, [[PILImage.create, Resize((35,28))], _pnt_lbl])
pnt_tdl = TfmdDL(pnt_dsrc.train, after_item=[PointScaler(), ToTensor()],
after_batch=[IntToFloatTensor(), *aug_transforms(max_warp=0)], bs=9)
pnt_tdl.show_batch(max_n=9)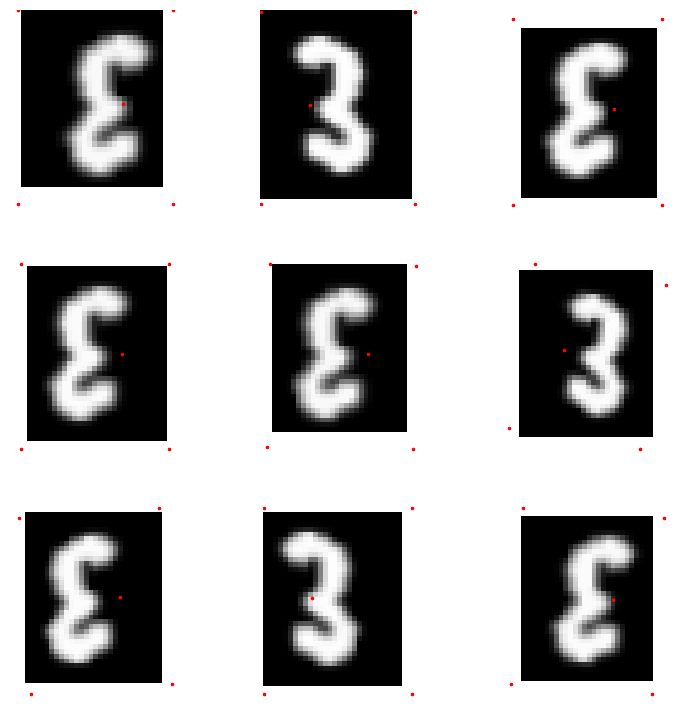
Bounding boxes
coco = untar_data(URLs.COCO_TINY)
images, lbl_bbox = get_annotations(coco/'train.json')
idx=2
coco_fn,bbox = coco/'train'/images[idx],lbl_bbox[idx]
def _coco_bb(x): return TensorBBox.create(bbox[0])
def _coco_lbl(x): return bbox[1]coco_dsrc = Datasets([coco_fn]*10, [PILImage.create, [_coco_bb], [_coco_lbl, MultiCategorize(add_na=True)]], n_inp=1)
coco_tdl = TfmdDL(coco_dsrc, bs=9, after_item=[BBoxLabeler(), PointScaler(), ToTensor(), Resize(256)],
after_batch=[IntToFloatTensor(), *aug_transforms()])
coco_tdl.show_batch(max_n=9)