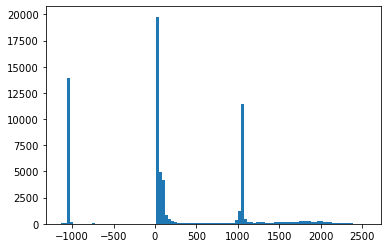
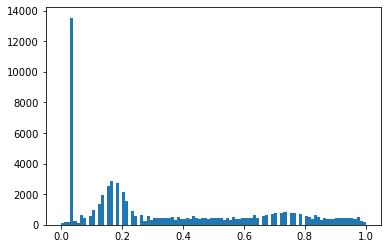
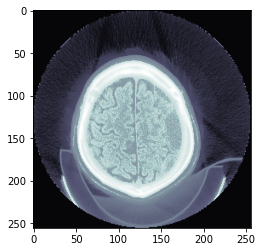
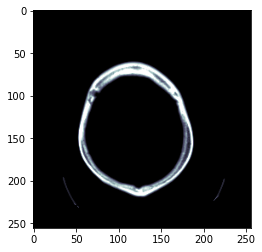
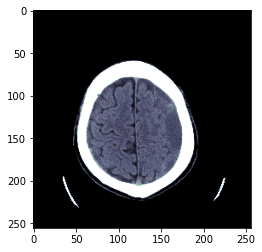
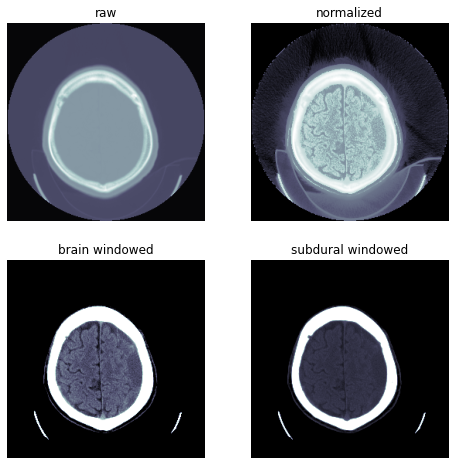
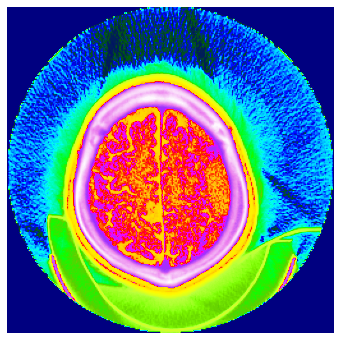
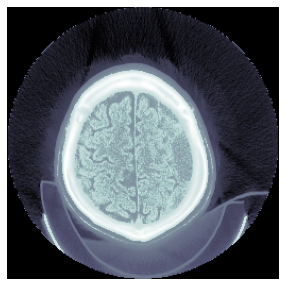
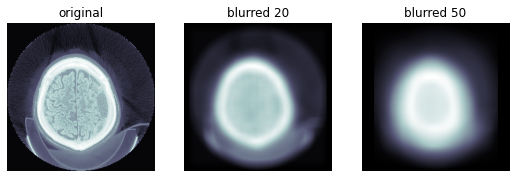
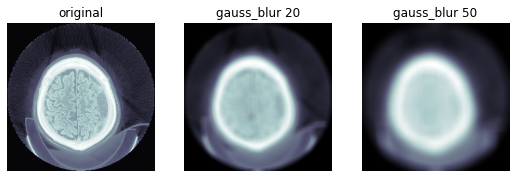
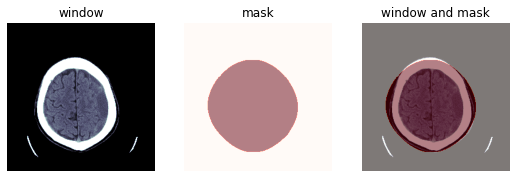
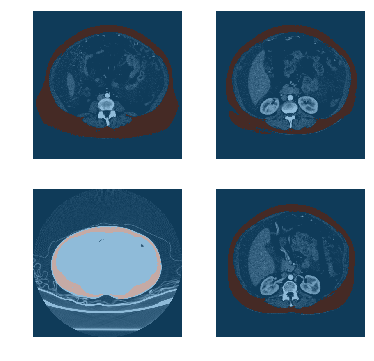
TEST_DCM = Path('images/sample.dcm')
dcm = TEST_DCM.dcmread()
dcmDataset.file_meta -------------------------------
(0002, 0000) File Meta Information Group Length UL: 176
(0002, 0001) File Meta Information Version OB: b'\x00\x01'
(0002, 0002) Media Storage SOP Class UID UI: CT Image Storage
(0002, 0003) Media Storage SOP Instance UID UI: 9999.180975792154576730321054399332994563536
(0002, 0010) Transfer Syntax UID UI: Explicit VR Little Endian
(0002, 0012) Implementation Class UID UI: 1.2.40.0.13.1.1.1
(0002, 0013) Implementation Version Name SH: 'dcm4che-1.4.38'
-------------------------------------------------
(0008, 0018) SOP Instance UID UI: ID_e0cc6a4b5
(0008, 0060) Modality CS: 'CT'
(0010, 0020) Patient ID LO: 'ID_a107dd7f'
(0020, 000d) Study Instance UID UI: ID_6468bdd34a
(0020, 000e) Series Instance UID UI: ID_4be303ae64
(0020, 0010) Study ID SH: ''
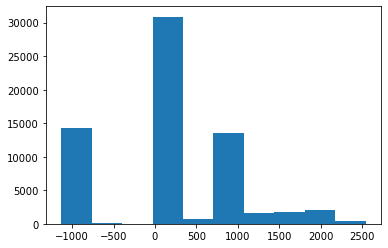
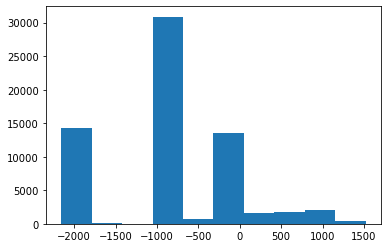
(0020, 0032) Image Position (Patient) DS: [-125.000, -122.268, 115.936]
(0020, 0037) Image Orientation (Patient) DS: [1.000000, 0.000000, 0.000000, 0.000000, 0.978148, -0.207912]
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0010) Rows US: 256
(0028, 0011) Columns US: 256
(0028, 0030) Pixel Spacing DS: [0.488281, 0.488281]
(0028, 0100) Bits Allocated US: 16
(0028, 0101) Bits Stored US: 16
(0028, 0102) High Bit US: 15
(0028, 0103) Pixel Representation US: 1
(0028, 1050) Window Center DS: "40.0"
(0028, 1051) Window Width DS: "100.0"
(0028, 1052) Rescale Intercept DS: "-1024.0"
(0028, 1053) Rescale Slope DS: "1.0"
(7fe0, 0010) Pixel Data OW: Array of 131072 elements